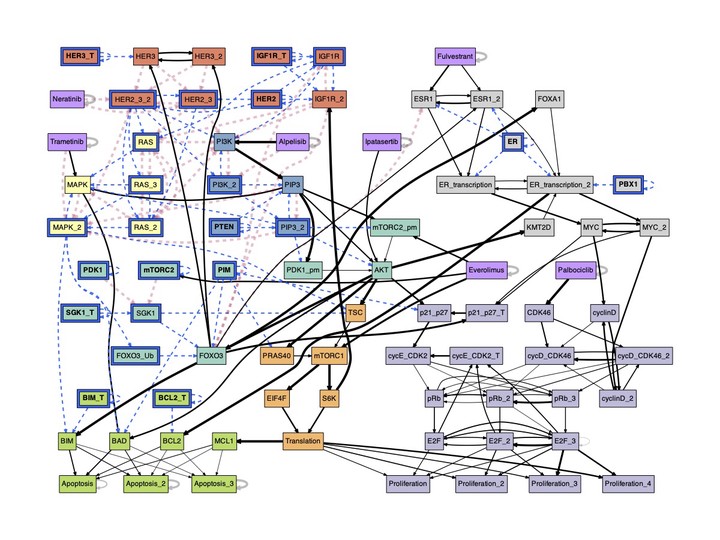
 The conditional effective graph of ER+ Breast Cancer.
The conditional effective graph of ER+ Breast Cancer.
Logical models offer a simple but powerful framework to understand the complex dynamics of biochemical regulation, without the need to estimate kinetic parameters. However, even simple automata components can lead to collective dynamics that are computationally intractable when aggregated into large networks.
Here we provide a publicly-available Python package that simulates the dynamics and control of Boolean Network models. CANA can also be used to calculate all measures of canalization that derive from removing dynamical redundancy via two-symbol schemata re-description (Marques-Pita and Rocha, 2013): effective connectivity, input redundancy, and input symmetry. At the network level, CANA also calculates the effective graph, a weighted and directed graph whose edge weights denote their effective contribution to node transitions. Finally, CANA can compute several measures of controllability that depend on the Controlled State Transition Graph.