Canalization & Control in Complex Systems
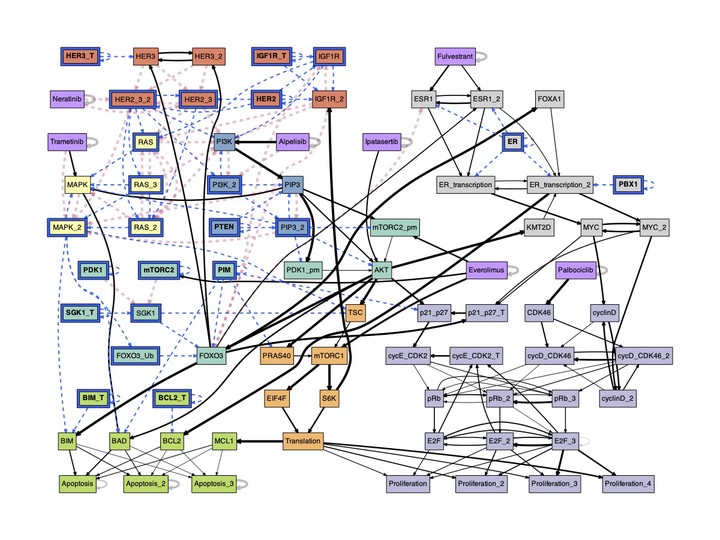
Many biological networks can be modeled with multivariate discrete dynamical systems. Current modeling theory suggests that the network structure of interactions captures salient features of system dynamics, but it misses a key aspect of these networks: some interactions are more important than others due to dynamical redundancy and nonlinearity. This unequivalence leads to a canalized dynamics that is different from dynamical constraints inferred from network structure alone.
To capture the redundancy present in biochemical regulatory and signaling interactions, we present the effective graph, an experimentally-validated mathematical framework that leads to a synthesis of both structure and dynamics in a new weighted graph representation of discrete multivariate systems. Our results demonstrate the ubiquity of redundancy in biology and provide a novel tool to increase causal explainability and control of biochemical regulation and signaling.
